What would you do with your microbiome sequence?
Alexandra Carmichael
August 1, 2012
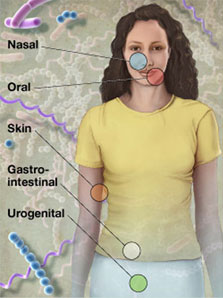
If you had access to free microbiome sequencing tests, to detect and analyze bacteria living in the nose, mouth, skin, gastro-intestinal, and/or urogenital areas of the body, what experiments would you think up?
Would you compare oral bacteria in people with lots of cavities vs. people with no cavities, look for differences between people with clear skin and acne, or sample your gut flora as you travel or change your diet? These are just examples — there are countless ideas.
As it turns out, we DO have up to 100 free microbiome profiles being offered to the QS community, thanks to Pathogenica and QS sponsor Autodesk. Now we just have to think up some cool experiments to do.
So the challenge is on – propose an experiment in the comments, and the top experiments will be done with some of the free tests. The deadline for submitting ideas is August 31. Also, all the data will be made openly available.
Check out some background reading on microbiome sequencing at the Human Microbiome Project website and Wikipedia.
Let’s come up with awesome ideas!